RStudio Snippets
productivity
Introduction
This tutorial demonstrates how to use both built-in and custom RStudio snippets within an R script. By leveraging snippets, you can streamline your coding process, reduce repetitive tasks, and maintain consistency across your projects.
Table of Contents
- Introduction to RStudio Snippets
- Using Built-in Snippets
- Creating and Using Custom Snippets
Introduction to RStudio Snippets
Snippets in RStudio are predefined blocks of code that can be quickly inserted into your scripts or markdown files. They enhance productivity by minimizing repetitive typing and ensuring consistency across your coding projects.
In this tutorial, we’ll explore how to utilize RStudio’s built-in snippets and create custom snippets tailored to your specific workflow.
Using Built-in Snippets
RStudio comes equipped with several built-in snippets that can be effortlessly used in .qmd files. Below are some commonly used built-in snippets along with examples of their usage.
Fun
Type fun and press Tab.
fun my_function <- function(arg1, arg2) {
# Function body
}For
Type for and press Tab.
for (variable in vector) {
}While
Type while and press Tab.
while (condition) {
}Apply
Type lapply and press Tab.
apply(array, margin, ...)Lapply
Type lapply and press Tab.
lapply(list, function)ts
Type ts and press Tab.
# Fri Oct 11 09:53:23 2024 ------------------------------Using Custom Snippets
Go to Tools > Global Options > Snippets > Edit.
Then paste the following at the bottom of the page and hit Apply
snippet dev
"nih-nci-dceg-connect-dev"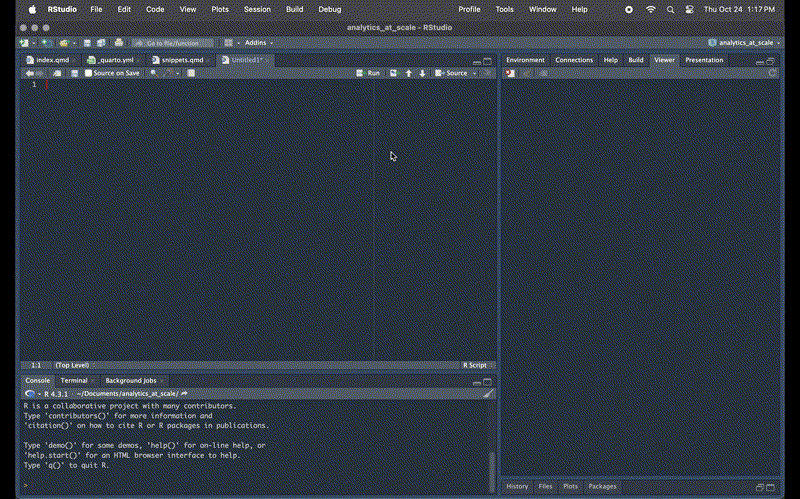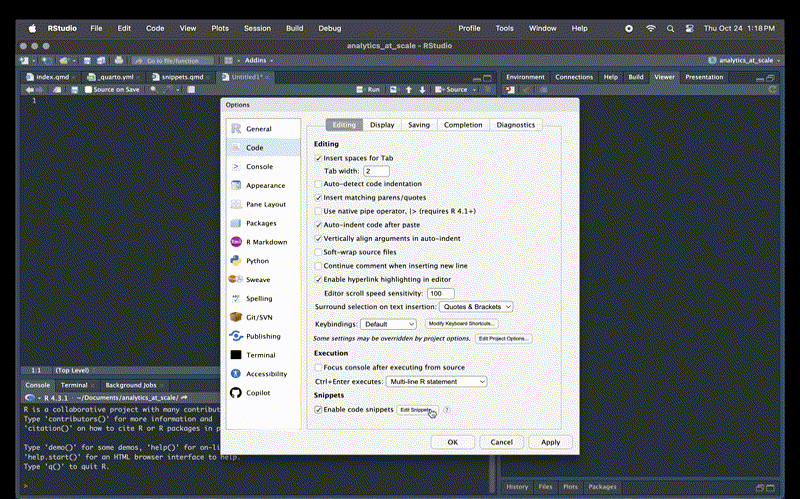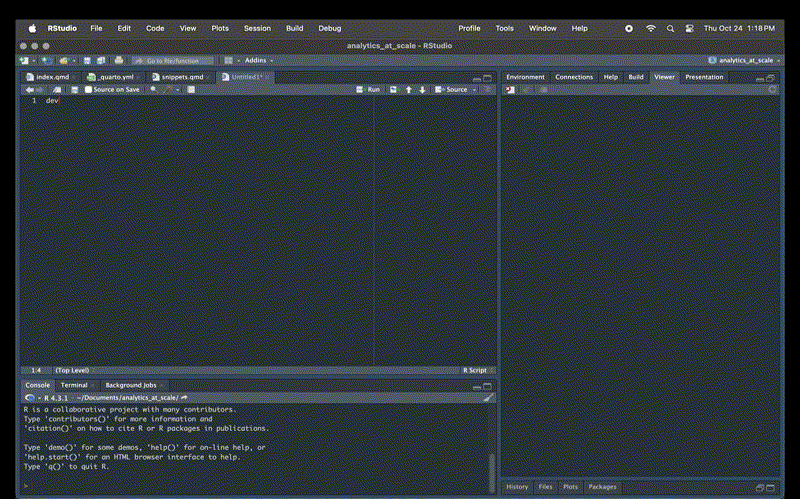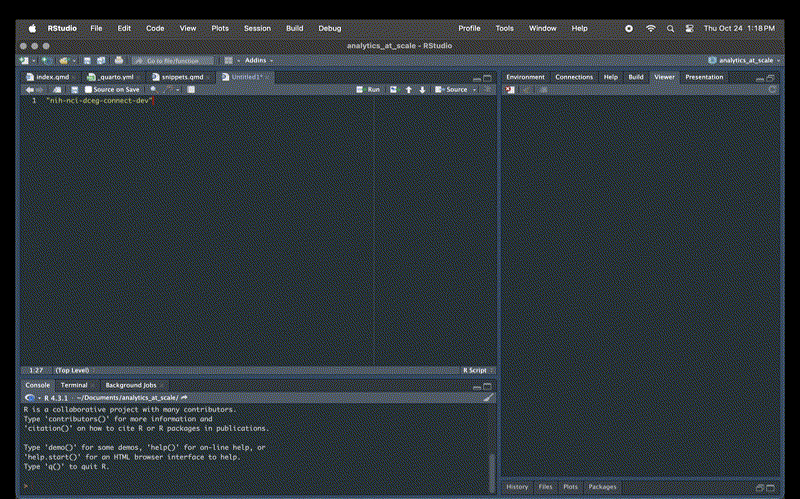
Note that all code after the snippet <name_of_snippet> key word must be indented.
More custom snippets to try
snippet header
## Description =================================================================
# Title: ${1:Script Title}
# Author: ${2:Your Name}
# Date: ${3:`r format(Sys.Date(), "%Y-%m-%d")`}
# Objective: ${4:Script Purpose}
# GH-Issue: ${5:Github Issue}
snippet query
bq_auth() # Authenticate with BigQuery
con <- DBI::dbConnect(bigrquery::bigquery(),
project=project, dataset=dataset, billing=billing))
# Specify just the data we need with a query
dataset <- "FlatConnect"
table <- "participants_JP"
tier <- "nih-nci-dceg-connect-prod-6d04"
sql <- glue::glue(
"""
SELECT
Connect_ID,
token
FROM `{project}.{dataset}.{table}`
""")
# Query the data and store as reference object
data <- dbGetQuery(con, SQL)
snippet dev
"nih-nci-dceg-connect-dev"
snippet stg
"nih-nci-dceg-connect-stg-5519"
snippet prod
"nih-nci-dceg-connect-prod-6d04"
snippet exclusions
WHERE
p.d_821247024 = '197316935' -- 'Verif Status' IS 'Verified'
AND p.d_747006172 != '353358909' -- 'Withdraw consent' IS NOT "Yes"
AND p.d_100767870 = '104430631' -- 'All Base Surv Complete' = 'No'
AND p.d_685002411_d_994064239 != '353358909' -- 'Ref Base Survs' IS NOT "Yes"
AND p.d_987563196 != '353358909' -- 'Dead' IS NOT 'Yes'